New COMPADRE & COMADRE versions are out!
by Rob Salguero-Gomez on Aug 11, 2016Yesterday, coinciding with the symposium "Landscape demography: Population dynamics across spatial scales" at the 2016 Ecological Society of America annual meeting, where we gave the talk "Global plant and animal demography: tearing the curtain and filling up the gap", we released the new two versions of the sister databases: COMPADRE version 4.0.0 and COMADRE version 2.0.0. These can be downloaded fully open-access at www.compadre-db.org. [caption id="attachment_media-6" align="alignnone" width="2142"]
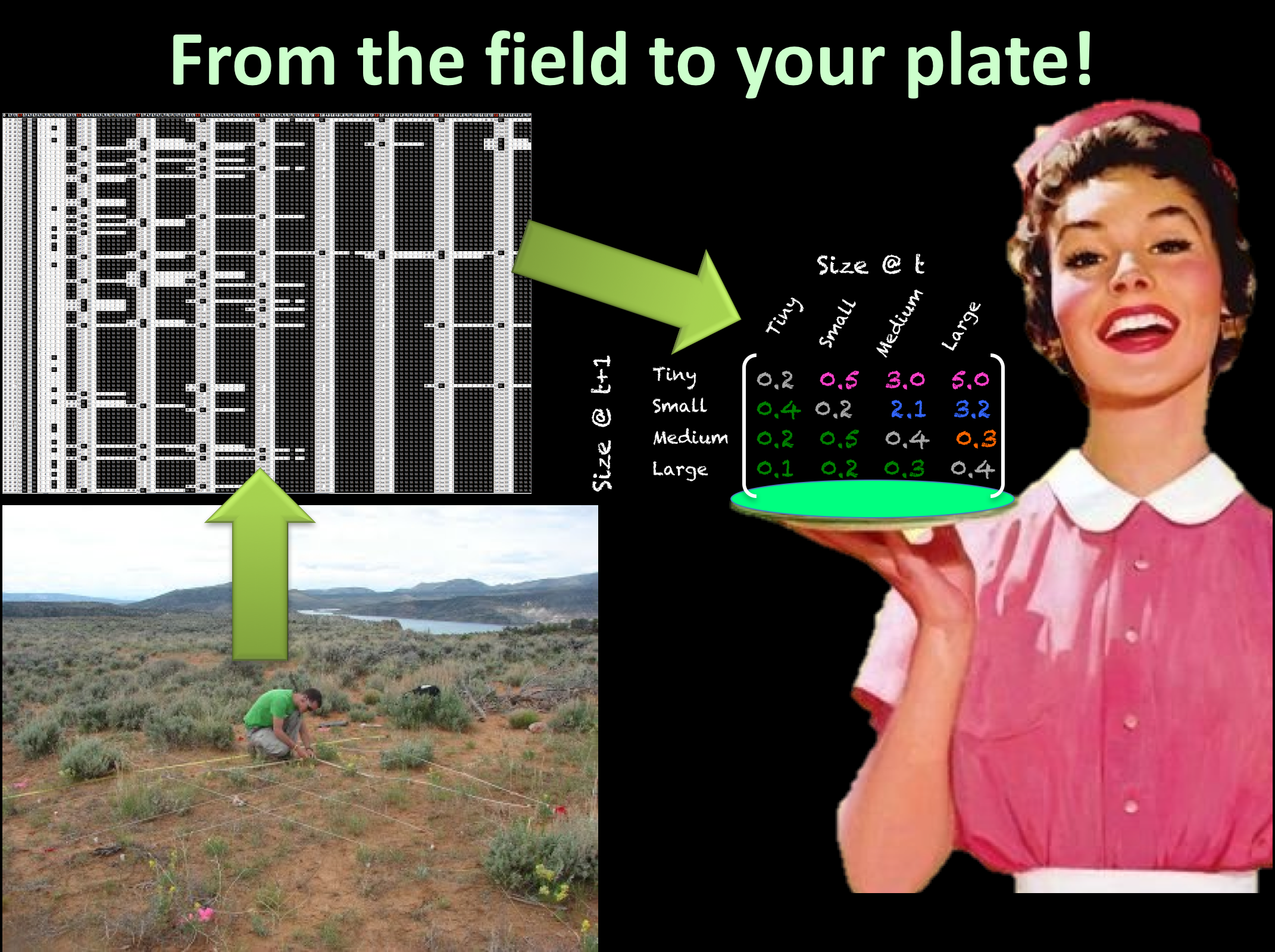 Our goals, at COMPADRE & COMADRE: to digitise and standardise matrix population models published and communicated to us by population ecologists, to error-check, fix and complement the information with metadata (e.g. taxonomy, phylogeny, biogeography), and to make it open access. In other words: to bring the field demographic data to your computer.[/caption]
What's new in them? More matrix population models, more species, more metadata! The COMPADRE Plant Matrix Database now contains 695 unique taxonomically accepted plant species from 819 published or personally communicated studies, adding up to a total of over 7,000 population matrix models. Similarly, the COMADRE Animal Matrix Database now contains 405 taxonomically accepted animal species outsourced from 508 studies, with a total of 1,927 matrix models.
[caption id="attachment_307" align="alignnone" width="2160"]
Our goals, at COMPADRE & COMADRE: to digitise and standardise matrix population models published and communicated to us by population ecologists, to error-check, fix and complement the information with metadata (e.g. taxonomy, phylogeny, biogeography), and to make it open access. In other words: to bring the field demographic data to your computer.[/caption]
What's new in them? More matrix population models, more species, more metadata! The COMPADRE Plant Matrix Database now contains 695 unique taxonomically accepted plant species from 819 published or personally communicated studies, adding up to a total of over 7,000 population matrix models. Similarly, the COMADRE Animal Matrix Database now contains 405 taxonomically accepted animal species outsourced from 508 studies, with a total of 1,927 matrix models.
[caption id="attachment_307" align="alignnone" width="2160"] Summary statistics of the new versions of COMPADRE and COMADRE: more species, more matrix population models, and more metadata.[/caption]
The increase in species and number matrices adds up to an unprecedented geographic cover. Clearly, however, geographic and taxonomic biases do exist in the databases, and we encourage users to carry out demographic research using matrix population models in under-explored areas (e.g. Belgium, Ireland, Italy, Greece, Russia, Morocco, Ecuador, Philippines, etc)
[caption id="attachment_315" align="alignnone" width="2168"]
Summary statistics of the new versions of COMPADRE and COMADRE: more species, more matrix population models, and more metadata.[/caption]
The increase in species and number matrices adds up to an unprecedented geographic cover. Clearly, however, geographic and taxonomic biases do exist in the databases, and we encourage users to carry out demographic research using matrix population models in under-explored areas (e.g. Belgium, Ireland, Italy, Greece, Russia, Morocco, Ecuador, Philippines, etc)
[caption id="attachment_315" align="alignnone" width="2168"]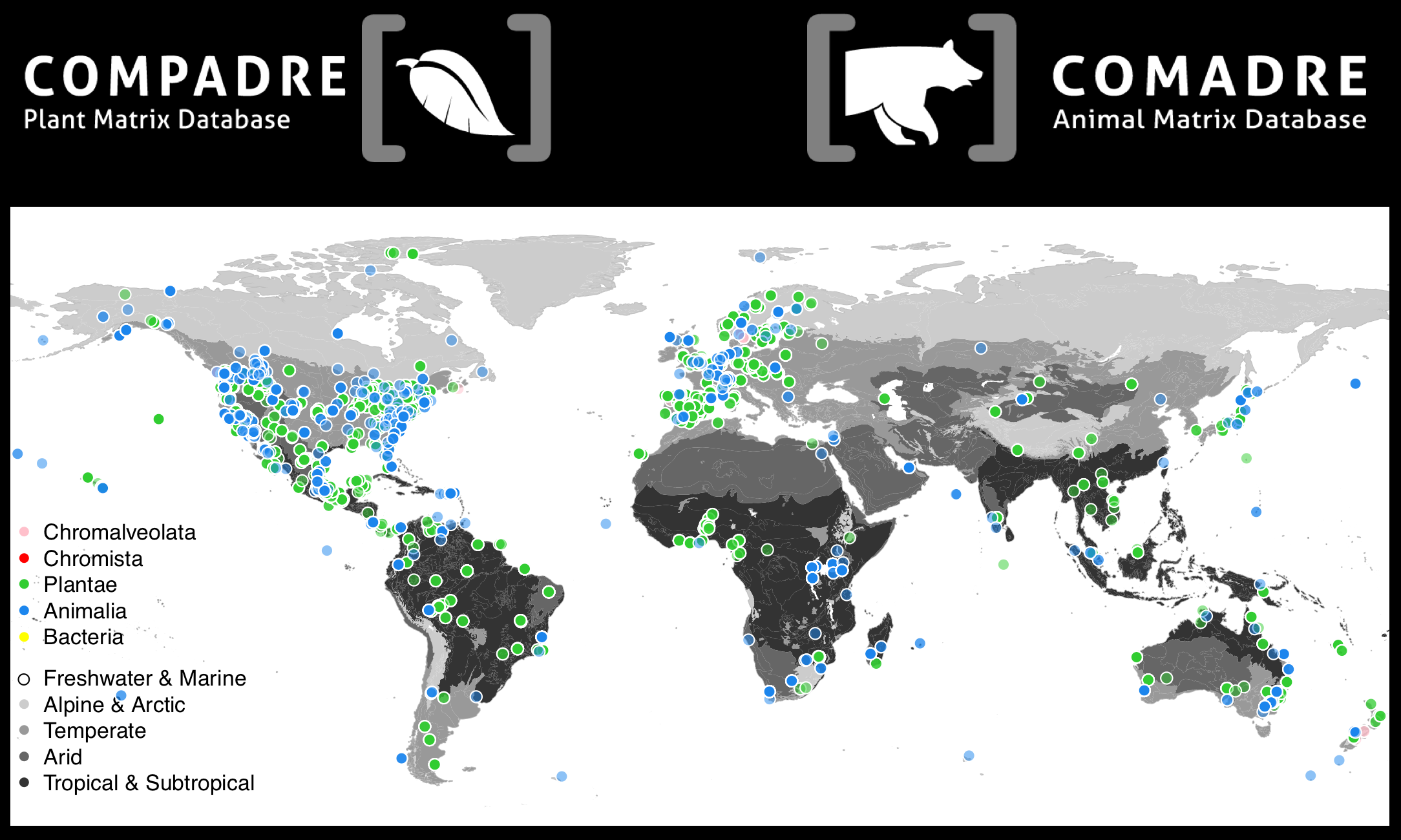 Geographic location of the studies where GPS information was provided in the publications, showing the global coverage of COMPADRE and COMADRE (Salguero-Gómez et al. in prep.)[/caption]
In addition to the new species and matrix models, we have also archived new variables. For instance, the latitudes and longitudes are no longer provided as separate degrees, minutes and seconds, but rather provided as two vectors (Lat and Lon), which is a more manageable format. For instance, try the following in your R console, after you have downloaded the Rdata objects and loaded them onto R:
Geographic location of the studies where GPS information was provided in the publications, showing the global coverage of COMPADRE and COMADRE (Salguero-Gómez et al. in prep.)[/caption]
In addition to the new species and matrix models, we have also archived new variables. For instance, the latitudes and longitudes are no longer provided as separate degrees, minutes and seconds, but rather provided as two vectors (Lat and Lon), which is a more manageable format. For instance, try the following in your R console, after you have downloaded the Rdata objects and loaded them onto R:
load("~/WhereverYouSaveYourStuff/COMPADRE_v.4.0.0.RData")
map("world",col="white",fill=T,bg="white",xlim=c(-175,176),ylim=c(-60,85),border="gray",mar=rep(0,4))
points(compadre$metadata[,c("Lon","Lat")])
We've also renamed some other variables to unify the organization of plants and animals, for instance:
load("~/WhereverYouSaveYourStuff/COMADRE_v.2.0.0.RData")
table(comadre$metadata$OrganismType)
load("~/WhereverYouSaveYourStuff/COMPADRE_v.4.0.0.RData")
table(compadre$metadata$OrganismType)
More information about the databases, their organisation, the way we digitise, complement, error-check and release information, and some useful workshop materials and R functions (incoming R package... stay tuned!) can be found here.
Happy COM(P)ADRE-ing!
The COMPADRE & COMADRE core committee
Rob Salguero-Gomez - The University of Sheffield & Max Planck Institute for Demographic Research (MPIDR)
Owen Jones - Southern Denmark University (SDU), MaxO
Ruth Archer - Exeter University
Yvonne Buckley - Trinity College Dublin
Judy Che-Castaldo - Lincoln Zoo
Hal Caswell - University of Amsterdam
Tom Ezard - University of Southampton
Dave Hodgson - Exeter University
Alex Scheuerlein - MPIDR
Jim Vaupel - MPIDR & SDU, MaxO


