A signature of life history in the stochastic dynamics of structured populations
by Rob Salguero-Gomez on Mar 4, 2017Different species have different average life histories. Such a variation is apparent from key species characteristics like longevity, age of maturity, or the number of offspring produced per clutch. Matrix population models such as those in the COMADRE Animal Matrix Database allow us to calculate and explore the diversity of such characteristics across species, and understand how they are connected. In a recent study published in Oikos, my colleague and I examined another key life history characteristic: demographic variance. This trait provides a measure of the total amount of stochasticity in a given life history arising from inherent randomness in survival, offspring production, and other individual-level demographic processes. Typically, species with a high demographic variance are short-lived and can produce many offspring per reproductive event, such as mice. Long-lived species, who typically produce only one offspring at a time, such as elephants, tend to have a much lower demographic variance. Using matrix population models from 24 mammal species from COMADRE, we calculated their demographic variance. We first considered how demographic variance is related to generation time (see Figure 1). As expected, there was a strong correlation with generation time, where species whose populations take longer to renew its individuals had a much lower demographic variance.
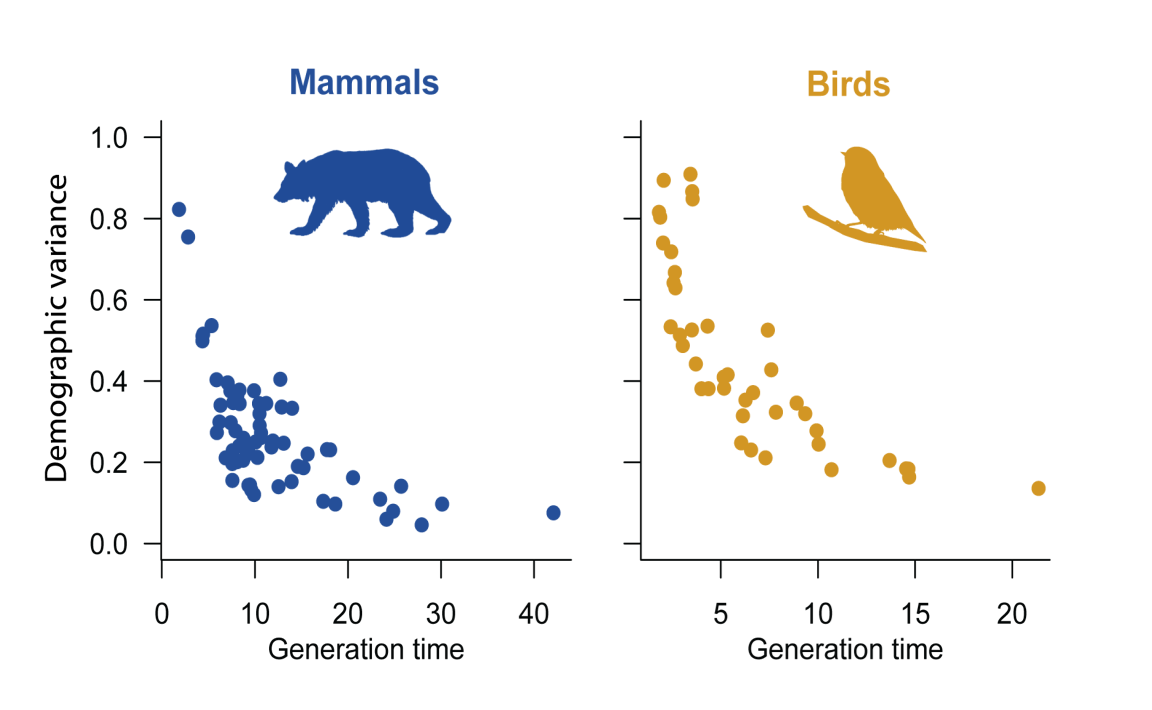 Figure 1. Demographic variance plotted against generation time, calculated using demographic information for mammals and birds from the COMADRE Animal Matrix Database.
Figure 1. Demographic variance plotted against generation time, calculated using demographic information for mammals and birds from the COMADRE Animal Matrix Database.
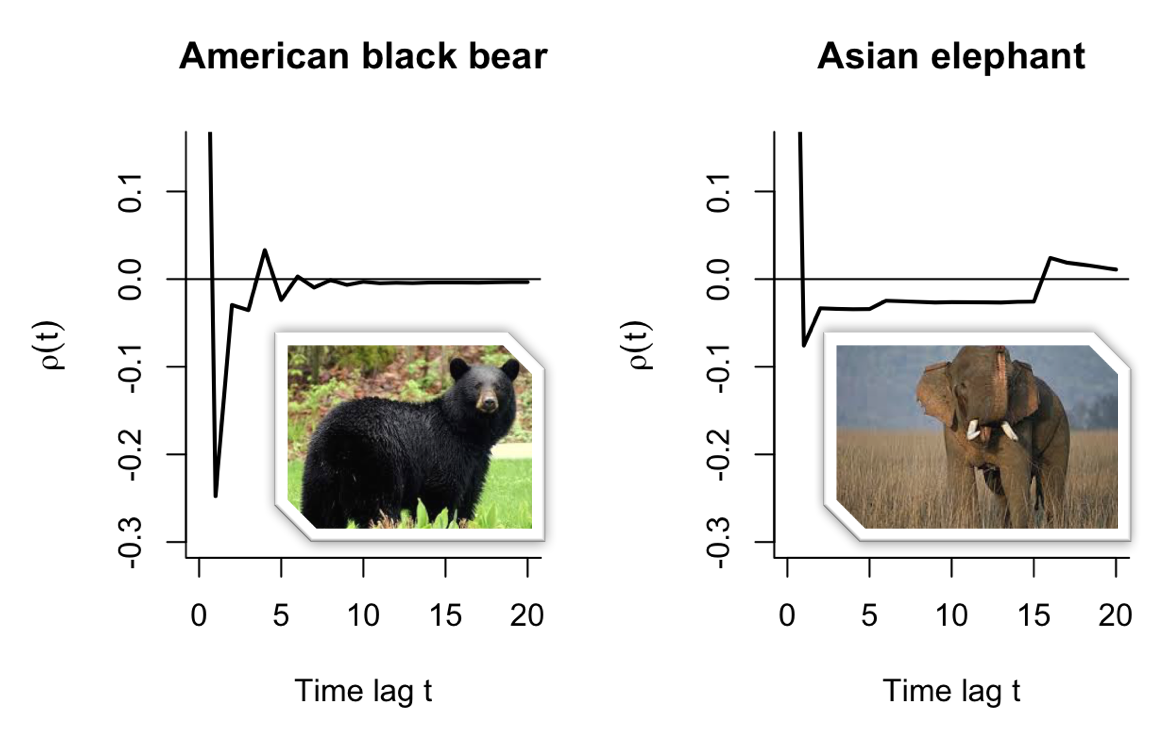
We then considered the temporal correlation in the stochastic population dynamics, which arise because of short-term fluctuations in the demographic structure. For each matrix model we estimated the autocorrelation function, which describes the degree to which two population growth increments tend to be similar for different time steps. This function is different for each model, and represents a signature of the life history and demographic stochasticity (see examples in Figure 2). A main result from our study is that the sum of these correlations over time lags describes the impact of population structure on the demographic variance.